Summary
Clan
This family is a member of clan (CL00050), which contains the following 2 members:
SNORD26 SNORD81Wikipedia annotation Edit Wikipedia article
The Rfam group coordinates the annotation of Rfam families in Wikipedia. This family is described by a Wikipedia entry Small nucleolar RNA SNORD81. More...
This page is based on a Wikipedia article. The text is available under the Creative Commons Attribution/Share-Alike License.
Sequences
Quick actions
Table preview (300 out of 339 sequence regions)
| Original order | Download FASTA | Accession | Bit score | Type | Start | End | Description | Species | View context |
|---|---|---|---|---|---|---|---|---|---|
| 0 | AJ224028.1 | N/A | seed | 3 | 79 | Homo sapiens Z23 small nucleolar RNA gene | Homo sapiens (human) |
|
|
| 1 | AJ224035.1 | N/A | seed | 1 | 76 | Mus musculus Z23 small nucleolar RNA gene | Mus musculus (house mouse) |
|
|
| 2 | AL606750.13 | N/A | seed | 40,184 | 40,260 | Mouse DNA sequence from clone RP23-335A19 on chromosome 3 Contains a glyceraldehyde-3-phosphate dehydrogenase (Gapd) pseudogene, the 3' end of two novel genes (IMAGE:6306854 IMAGE:5030542 4930431B09Rik) and a novel gene. | Mus musculus (house mouse) |
|
|
| 3 | ABDC01278790.1 | N/A | seed | 3,802 | 3,878 | Microcebus murinus cont1.278789, whole genome shotgun sequence. | Microcebus murinus (gray mouse lemur) |
|
|
| 4 | AANU01240317.1 | N/A | seed | 3,294 | 3,218 | Macaca mulatta 1099214671913, whole genome shotgun sequence. | Macaca mulatta (Rhesus monkey) |
|
|
| 5 | AAPY01728088.1 | N/A | seed | 9,340 | 9,415 | Tupaia belangeri cont1.728087, whole genome shotgun sequence. | Tupaia belangeri (northern tree shrew) |
|
|
| 6 | AAYZ01154866.1 | N/A | seed | 3,472 | 3,396 | Ochotona princeps cont2.154865, whole genome shotgun sequence. | Ochotona princeps (American pika) |
|
|
| 7 | AALT01063557.1 | N/A | seed | 160 | 231 | Sorex araneus cont1.063556, whole genome shotgun sequence. | Sorex araneus (European shrew) |
|
|
| 8 | AAFC03120161.1 | N/A | seed | 30,906 | 30,982 | Bos taurus Ctg33835, whole genome shotgun sequence. | Bos taurus (cattle) |
|
|
| 9 | AADA01033599.1 | N/A | seed | 12,200 | 12,125 | Pan troglodytes ctg_33598, whole genome shotgun sequence. | Pan troglodytes (chimpanzee) |
|
|
| 10 | AANN01159637.1 | N/A | seed | 259 | 333 | Erinaceus europaeus cont1.159636, whole genome shotgun sequence. | Erinaceus europaeus (western European hedgehog) |
|
|
| 11 | AAIY01534871.1 | N/A | seed | 472 | 399 | Echinops telfairi cont1.534871, whole genome shotgun sequence. | Echinops telfairi (small Madagascar hedgehog) |
|
|
| 12 | AANU01265424.1 | N/A | seed | 25,442 | 25,518 | Macaca mulatta 1099214598121, whole genome shotgun sequence. | Macaca mulatta (Rhesus monkey) |
|
|
| 13 | AAIY01210190.1 | N/A | seed | 1,318 | 1,402 | Echinops telfairi cont1.210189, whole genome shotgun sequence. | Echinops telfairi (small Madagascar hedgehog) |
|
|
| 14 | AAIY01244248.1 | N/A | seed | 1,113 | 1,040 | Echinops telfairi cont1.244247, whole genome shotgun sequence. | Echinops telfairi (small Madagascar hedgehog) |
|
|
| 15 | AANU01202269.1 | N/A | seed | 48,062 | 48,138 | Macaca mulatta 1099214710582, whole genome shotgun sequence. | Macaca mulatta (Rhesus monkey) |
|
|
| 16 | AAGV020273689.1 | N/A | seed | 3,634 | 3,710 | Dasypus novemcinctus cont2.273688, whole genome shotgun sequence. | Dasypus novemcinctus (nine-banded armadillo) |
|
|
| 17 | AAKN02037311.1 | N/A | seed | 43,777 | 43,853 | Cavia porcellus strain inbred line 2N cont2.37310, whole genome shotgun sequence. | Cavia porcellus (Domestic guinea pig) |
|
|
| 18 | AAGU03036956.1 | N/A | seed | 70,179 | 70,252 | Loxodonta africana cont3.36955, whole genome shotgun sequence. | Loxodonta africana (African savanna elephant) |
|
|
| 19 | AAEX03005203.1 | N/A | seed | 132,526 | 132,602 | Canis lupus familiaris cont3.5203, whole genome shotgun sequence. | Canis lupus familiaris (dog) |
|
|
| 20 | AAPE02018271.1 | N/A | seed | 28,237 | 28,161 | Myotis lucifugus cont2.18270, whole genome shotgun sequence. | Myotis lucifugus (little brown bat) |
|
|
| 21 | AAWZ02040398.1 | N/A | seed | 3,978 | 3,901 | Anolis carolinensis chromosome Unknown cont2.40397, whole genome shotgun sequence. | Anolis carolinensis (green anole) |
|
|
| 0 | KZ206965.1 | 107.80 | full | 21,208,115 | 21,208,191 | KZ206965.1 Aotus nancymaae isolate 86115 unplaced genomic scaffold Scaffold76, whole genome shotgun sequence | Aotus nancymaae (Ma's night monkey) |
|
|
| 1 | JACDTQ010002688.1 | 107.80 | full | 10,134,704 | 10,134,628 | JACDTQ010002688.1 Diceros bicornis minor isolate SBR-YM ScS9zPn_2688, whole genome shotgun sequence | Diceros bicornis minor |
|
|
| 2 | NW_017731356.1 | 107.80 | full | 4,780,640 | 4,780,564 | NW_017731356.1 Hipposideros armiger isolate ML-2016 unplaced genomic scaffold, ASM189008v1, whole genome shotgun sequence | Hipposideros armiger |
|
|
| 3 | CM001658.1 | 107.10 | full | 28,653,665 | 28,653,741 | Nomascus leucogenys chromosome 12, whole genome shotgun sequence. | Nomascus leucogenys (Northern white-cheeked gibbon) |
|
|
| 4 | CM009262.2 | 107.10 | full | 76,329,182 | 76,329,258 | CM009262.2 Pongo abelii isolate Susie chromosome 1, whole genome shotgun sequence | Pongo abelii (Sumatran orangutan) |
|
|
| 5 | FR853080.2 | 107.10 | full | 152,936,286 | 152,936,210 | Gorilla gorilla gorilla genomic chromosome, chr1, whole genome shotgun sequence | Gorilla gorilla gorilla (western lowland gorilla) |
|
|
| 6 | CM021932.1 | 106.20 | full | 22,742,984 | 22,743,060 | CM021932.1 Callithrix jacchus isolate mCalJac1 chromosome 18, whole genome shotgun sequence | Callithrix jacchus (white-tufted-ear marmoset) |
|
|
| 7 | NW_017869562.1 | 105.90 | full | 2,083,524 | 2,083,448 | NW_017869562.1 Castor canadensis isolate Ward unplaced genomic scaffold, C.can genome v1.0 scaffold_235, whole genome shotgun sequence | Castor canadensis (American beaver) |
|
|
| 8 | CAAGRJ010041540.1 | 105.40 | full | 219,352 | 219,428 | CAAGRJ010041540.1 Lynx pardinus genome assembly, contig: lp23s15824, whole genome shotgun sequence | Lynx pardinus (Spanish lynx) |
|
|
| 9 | CM001394.3 | 105.40 | full | 18,038,019 | 18,037,943 | Felis catus isolate Cinnamon breed Abyssinian chromosome F1, whole genome shotgun sequence. | Felis catus (domestic cat) |
|
|
| 10 | CM017345.2 | 105.40 | full | 15,787,069 | 15,786,993 | CM017345.2 Lynx canadensis isolate LIC74 chromosome F1, whole genome shotgun sequence | Lynx canadensis (Canada lynx) |
|
|
| 11 | LZNR01001545.1 | 105.40 | full | 216,954 | 216,878 | LZNR01001545.1 Ursus americanus isolate MABB319796 Contig1547, whole genome shotgun sequence | Ursus americanus (American black bear) |
|
|
| 12 | NW_020339222.1 | 105.40 | full | 284,521 | 284,445 | NW_020339222.1 Puma concolor isolate SC36_Marlon unplaced genomic scaffold, PumCon1.0 scaffold_2175, whole genome shotgun sequence | Puma concolor (puma) |
|
|
| 13 | NW_020834759.1 | 105.40 | full | 229,716 | 229,640 | NW_020834759.1 Acinonyx jubatus isolate Rico unplaced genomic scaffold, Aci_jub_2 Primary Assembly Aci_jub_2_ph1_scaffold33, whole genome shotgun sequence | Acinonyx jubatus (cheetah) |
|
|
| 14 | NW_024424452.1 | 105.40 | full | 77,697,044 | 77,696,968 | NW_024424452.1 Ursus maritimus isolate PB19 unplaced genomic scaffold, ASM1731132v1 scaffold_2, whole genome shotgun sequence | Ursus maritimus (polar bear) |
|
|
| 15 | CM016647.2 | 105.20 | full | 9,487,366 | 9,487,442 | CM016647.2 Camelus dromedarius isolate Drom800 breed African chromosome 21, whole genome shotgun sequence | Camelus dromedarius (Arabian camel) |
|
|
| 16 | KB016345.1 | 105.20 | full | 621,974 | 622,050 | Camelus ferus unplaced genomic scaffold scaffold45, whole genome shotgun sequence. | Camelus ferus (Wild Bactrian camel) |
|
|
| 17 | NW_011513524.1 | 105.20 | full | 191,102 | 191,026 | NW_011513524.1 Camelus bactrianus breed Alxa unplaced genomic scaffold, Ca_bactrianus_MBC_1.0 scaffold117, whole genome shotgun sequence | Camelus bactrianus (Bactrian camel) |
|
|
| 18 | NW_021964199.1 | 105.20 | full | 8,937,308 | 8,937,232 | NW_021964199.1 Vicugna pacos isolate Carlotta (AHFN-0088) chromosome 21 unlocalized genomic scaffold, VicPac3.1 ScfyRBE_283;HRSCAF=923, whole genome shotgun sequence | Vicugna pacos (alpaca) |
|
|
| 19 | KV389488.1 | 104.90 | full | 3,132,642 | 3,132,718 | KV389488.1 Cebus capucinus imitator isolate Cc_AM_T3 unplaced genomic scaffold Scaffold62, whole genome shotgun sequence | Cebus imitator (Panamanian white-faced capuchin) |
|
|
| 20 | NW_022436965.1 | 104.90 | full | 16,437,466 | 16,437,542 | NW_022436965.1 Sapajus apella isolate SASKATOON/1434 unplaced genomic scaffold, GSC_monkey_1.0 scaffold25, whole genome shotgun sequence | Cebus apella (Tufted capuchin) |
|
|
| 21 | CM014247.1 | 104.70 | full | 8,725,490 | 8,725,414 | CM014247.1 Rhinolophus ferrumequinum isolate MPI-CBG mRhiFer1 chromosome 22, whole genome shotgun sequence | Rhinolophus ferrumequinum (greater horseshoe bat) |
|
|
| 22 | CM007662.1 | 104.70 | full | 135,005,696 | 135,005,620 | CM007662.1 Microcebus murinus isolate mixed chromosome 2, whole genome shotgun sequence | Microcebus murinus (gray mouse lemur) |
|
|
| 23 | CM000314.3 | 104.10 | full | 152,270,715 | 152,270,639 | Pan troglodytes isolate Yerkes chimp pedigree #C0471 (Clint) chromosome 1, whole genome shotgun sequence. | Pan troglodytes (chimpanzee) |
|
|
| 24 | CM000663.2 | 104.10 | full | 173,864,222 | 173,864,146 | Homo sapiens chromosome 1, GRCh38 reference primary assembly. | Homo sapiens (human) |
|
|
| 25 | CM003383.1 | 104.10 | full | 153,086,193 | 153,086,117 | CM003383.1 Pan paniscus isolate Ulindi chromosome 1, whole genome shotgun sequence | Pan paniscus (pygmy chimpanzee) |
|
|
| 26 | CM026986.1 | 103.70 | full | 73,306,700 | 73,306,776 | CM026986.1 Rattus norvegicus strain BN/NHsdMcwi chromosome 13, whole genome shotgun sequence | Rattus norvegicus (Norway rat) |
|
|
| 27 | GL873535.1 | 103.50 | full | 19,291,269 | 19,291,345 | Otolemur garnettii unplaced genomic scaffold scaffold00016, whole genome shotgun sequence. | Otolemur garnettii (small-eared galago) |
|
|
| 28 | NW_019154108.1 | 103.20 | full | 28,346,463 | 28,346,539 | NW_019154108.1 Enhydra lutris kenyoni isolate GAN:26980312 unplaced genomic scaffold, ASM228890v2 scaffoldscaffold24, whole genome shotgun sequence | Enhydra lutris kenyoni |
|
|
| 29 | LZPO01034931.1 | 102.80 | full | 27,745 | 27,821 | Neotoma lepida isolate 417 scaffold_1122, whole genome shotgun sequence. | Neotoma lepida (desert woodrat) |
|
|
| 30 | NW_006501159.1 | 102.80 | full | 903,582 | 903,506 | NW_006501159.1 Peromyscus maniculatus bairdii unplaced genomic scaffold, Pman_1.0 Scaffold123, whole genome shotgun sequence | Peromyscus maniculatus bairdii (prairie deer mouse) |
|
|
| 31 | CM014336.1 | 102.80 | full | 31,163,526 | 31,163,450 | CM014336.1 Macaca mulatta isolate AG07107 chromosome 1, whole genome shotgun sequence | Macaca mulatta (Rhesus monkey) |
|
|
| 32 | CM018180.1 | 102.80 | full | 187,997,131 | 187,997,207 | CM018180.1 Papio anubis isolate 15944 chromosome 1, whole genome shotgun sequence | Papio anubis (Olive baboon) |
|
|
| 33 | CM019240.1 | 102.80 | full | 30,511,252 | 30,511,176 | CM019240.1 Piliocolobus tephrosceles isolate RC106 chromosome 1, whole genome shotgun sequence | Piliocolobus tephrosceles (Ugandan red Colobus) |
|
|
| 34 | KN296157.1 | 102.80 | full | 1,054,804 | 1,054,728 | KN296157.1 Rhinopithecus roxellana isolate Xiao Hai unplaced genomic scaffold ENSRROG022268, whole genome shotgun sequence | Rhinopithecus roxellana (golden snub-nosed monkey) |
|
|
| 35 | KN974384.1 | 102.80 | full | 187,361 | 187,285 | KN974384.1 Mandrillus leucophaeus isolate KB7577 unplaced genomic scaffold Scaffold226, whole genome shotgun sequence | Mandrillus leucophaeus (drill) |
|
|
| 36 | KN984673.1 | 102.80 | full | 4,437,927 | 4,437,851 | KN984673.1 Colobus angolensis palliatus isolate OR3802 unplaced genomic scaffold Scaffold59, whole genome shotgun sequence | Colobus angolensis palliatus |
|
|
| 37 | KQ004830.1 | 102.80 | full | 3,480,321 | 3,480,245 | KQ004830.1 Macaca nemestrina isolate M95218 unplaced genomic scaffold Scaffold12, whole genome shotgun sequence | Macaca nemestrina (pig-tailed macaque) |
|
|
| 38 | KQ010861.1 | 102.80 | full | 2,277,807 | 2,277,731 | KQ010861.1 Cercocebus atys isolate FAK unplaced genomic scaffold Scaffold232, whole genome shotgun sequence | Cercocebus atys (sooty mangabey) |
|
|
| 39 | MCGX01001328.1 | 102.80 | full | 2,962,927 | 2,962,851 | MCGX01001328.1 Rhinopithecus bieti isolate Rb0 scaffold_1853, whole genome shotgun sequence | Rhinopithecus bieti (Black snub-nosed monkey) |
|
|
| 40 | GL429821.1 | 102.60 | full | 1,168,103 | 1,168,027 | Myotis lucifugus unplaced genomic scaffold scaffold_54, whole genome shotgun sequence. | Myotis lucifugus (little brown bat) |
|
|
| 41 | JAULJE010000022.1 | 102.60 | full | 19,334,404 | 19,334,328 | JAULJE010000022.1 Eptesicus nilssonii isolate BLF_Eptnil Scaffold_22, whole genome shotgun sequence | Eptesicus nilssoni (northern bat) |
|
|
| 42 | KE162497.1 | 102.60 | full | 5,676,456 | 5,676,380 | KE162497.1 Myotis brandtii unplaced genomic scaffold scaffold250, whole genome shotgun sequence | Myotis brandtii |
|
|
| 43 | JH378117.1 | 102.60 | full | 13,721,707 | 13,721,783 | JH378117.1 Saimiri boliviensis boliviensis unplaced genomic scaffold scaffold00013, whole genome shotgun sequence | Saimiri boliviensis boliviensis (Bolivian squirrel monkey) |
|
|
| 44 | JH378118.1 | 102.60 | full | 33,676,544 | 33,676,620 | JH378118.1 Saimiri boliviensis boliviensis unplaced genomic scaffold scaffold00014, whole genome shotgun sequence | Saimiri boliviensis boliviensis (Bolivian squirrel monkey) |
|
|
| 45 | CM001966.2 | 102.00 | full | 55,327,842 | 55,327,918 | Chlorocebus sabaeus isolate 1994-021 chromosome 25, whole genome shotgun sequence. | Chlorocebus sabaeus |
|
|
| 46 | NW_004801631.1 | 102.00 | full | 9,966,233 | 9,966,309 | NW_004801631.1 Mesocricetus auratus isolate Golden Hamster female 1 unplaced genomic scaffold, MesAur1.0 scaffold00028, whole genome shotgun sequence | Mesocricetus auratus (golden hamster) |
|
|
| 47 | JACASE010000017.1 | 101.90 | full | 18,830,970 | 18,831,046 | JACASE010000017.1 Rousettus aegyptiacus isolate mRouAeg1 scaffold_m13_p_17, whole genome shotgun sequence | Rousettus aegyptiacus (Egyptian rousette) |
|
|
| 48 | JAATJV010186628.1 | 101.70 | full | 92,730 | 92,654 | JAATJV010186628.1 Sciurus carolinensis isolate SUZIE scaffold14528, whole genome shotgun sequence | Sciurus carolinensis (gray squirrel) |
|
|
| 49 | NW_025422087.1 | 101.60 | full | 15,970,014 | 15,969,938 | NW_025422087.1 Mustela putorius furo isolate JIRA1106 unplaced genomic scaffold, ASM1176430v1.1 tig00003942_pilon_pilon_pilon, whole genome shotgun sequence | Mustela putorius furo (domestic ferret) |
|
|
| 50 | CM025106.1 | 101.30 | full | 25,116,610 | 25,116,686 | CM025106.1 Canis lupus familiaris isolate SID07034 breed Labrador retriever chromosome 7, whole genome shotgun sequence | Canis lupus familiaris (dog) |
|
|
| 51 | CM025463.1 | 101.30 | full | 25,067,075 | 25,067,151 | CM025463.1 Canis lupus dingo isolate Sandy chromosome 7, whole genome shotgun sequence | Canis lupus dingo (dingo) |
|
|
| 52 | NW_020356470.1 | 101.30 | full | 15,085,473 | 15,085,549 | NW_020356470.1 Vulpes vulpes strain TameXAggressive cross unplaced genomic scaffold, VulVul2.2 scaffold36, whole genome shotgun sequence | Vulpes vulpes (red fox) |
|
|
| 53 | KQ025513.1 | 101.10 | full | 6,913,264 | 6,913,188 | KQ025513.1 Propithecus coquereli isolate 6110/MARCELLA unplaced genomic scaffold Scaffold56, whole genome shotgun sequence | Propithecus coquereli (Coquerel's sifaka) |
|
|
| 54 | NW_004450683.1 | 100.30 | full | 746,479 | 746,555 | NW_004450683.1 Odobenus rosmarus divergens isolate Ivan unplaced genomic scaffold, Oros_1.0 Scaffold420, whole genome shotgun sequence | Odobenus rosmarus divergens (Pacific walrus) |
|
|
| 55 | NW_020312885.1 | 100.30 | full | 17,253,202 | 17,253,126 | NW_020312885.1 Callorhinus ursinus isolate GAN/ISIS: 27379988/994484 unplaced genomic scaffold, ASM326570v1 scaffold143, whole genome shotgun sequence | Callorhinus ursinus (northern fur seal) |
|
|
| 56 | NW_017871545.1 | 100.20 | full | 14,028 | 14,104 | NW_017871545.1 Castor canadensis isolate Ward unplaced genomic scaffold, C.can genome v1.0 scaffold_2218, whole genome shotgun sequence | Castor canadensis (American beaver) |
|
|
| 57 | KB320813.1 | 100.10 | full | 1,638,627 | 1,638,702 | KB320813.1 Tupaia chinensis unplaced genomic scaffold Scaffold000369_1, whole genome shotgun sequence | Tupaia chinensis (Chinese tree shrew) |
|
|
| 58 | JACAGB010000018.1 | 100.10 | full | 21,776,515 | 21,776,591 | JACAGB010000018.1 Pipistrellus kuhlii isolate mPipKuh1 scaffold_m20_p_18, whole genome shotgun sequence | Pipistrellus kuhlii (Kuhl's pipistrelle) |
|
|
| 59 | JH378133.1 | 100.00 | full | 716,289 | 716,365 | JH378133.1 Saimiri boliviensis boliviensis unplaced genomic scaffold scaffold00029, whole genome shotgun sequence | Saimiri boliviensis boliviensis (Bolivian squirrel monkey) |
|
|
| 60 | JABWUV010000018.1 | 99.90 | full | 24,689,659 | 24,689,735 | JABWUV010000018.1 Myotis myotis isolate mMyoMyo1 scaffold_m19_p_18, whole genome shotgun sequence | Myotis myotis |
|
|
| 61 | KB109864.1 | 99.90 | full | 2,247,260 | 2,247,184 | KB109864.1 Myotis davidii unplaced genomic scaffold scaffold580, whole genome shotgun sequence | Myotis davidii |
|
|
| 62 | CM021939.1 | 99.80 | full | 31,127,195 | 31,127,119 | CM021939.1 Macaca fascicularis isolate cy0333 breed Mauritian cynomolgus macaque chromosome 1, whole genome shotgun sequence | Macaca fascicularis (crab-eating macaque) |
|
|
| 63 | CM018477.1 | 99.70 | full | 14,521,735 | 14,521,659 | CM018477.1 Panthera leo isolate Brooke chromosome F2, whole genome shotgun sequence | Panthera leo (lion) |
|
|
| 64 | CM000802.1 | 99.60 | full | 1,208,759 | 1,208,683 | Oryctolagus cuniculus chromosome 13, whole genome shotgun sequence. | Oryctolagus cuniculus (rabbit) |
|
|
| 65 | NC_058408.1 | 99.60 | full | 52,658,762 | 52,658,838 | NC_058408.1 Neomonachus schauinslandi chromosome 6, ASM220157v2, whole genome shotgun sequence | Monachus schauinslandi (Hawaiian monk seal) |
|
|
| 66 | NW_006383638.1 | 99.60 | full | 120,241 | 120,317 | NW_006383638.1 Leptonychotes weddellii isolate WS11-02 unplaced genomic scaffold, LepWed1.0 scaffold00598, whole genome shotgun sequence | Leptonychotes weddellii (Weddell seal) |
|
|
| 67 | CM000994.3 | 99.40 | full | 160,865,878 | 160,865,953 | CM000994.3 Mus musculus chromosome 1, GRCm39 reference primary assembly C57BL/6J | Mus musculus (house mouse) |
|
|
| 68 | QGOO02000001.1 | 99.40 | full | 34,519,236 | 34,519,161 | QGOO02000001.1 Mus spicilegus strain ZRU musp714_scaffold_00001b, whole genome shotgun sequence | Mus spicilegus (steppe mouse) |
|
|
| 69 | CM009950.1 | 99.10 | full | 192,179,467 | 192,179,543 | CM009950.1 Theropithecus gelada isolate Dixy chromosome 1, whole genome shotgun sequence | Theropithecus gelada (gelada baboon) |
|
|
| 70 | CM016973.1 | 99.00 | full | 124,034,529 | 124,034,452 | CM016973.1 Suricata suricatta isolate VVHF042 chromosome 3, whole genome shotgun sequence | Suricata suricatta (meerkat) |
|
|
| 71 | JH166641.1 | 98.90 | full | 1,885,577 | 1,885,653 | JH166641.1 Heterocephalus glaber unplaced genomic scaffold scaffold961, whole genome shotgun sequence | Heterocephalus glaber (naked mole-rat) |
|
|
| 72 | KE721788.1 | 98.80 | full | 2,955,998 | 2,956,074 | KE721788.1 Panthera tigris altaica isolate TaeGuk unplaced genomic scaffold scaffold177, whole genome shotgun sequence | Panthera tigris altaica (Amur tiger) |
|
|
| 73 | CABDUW010000713.1 | 98.50 | full | 693,145 | 693,068 | CABDUW010000713.1 Marmota monax genome assembly, contig: monax5_s00713, whole genome shotgun sequence | Marmota monax (woodchuck) |
|
|
| 74 | CZRN02000178.1 | 98.50 | full | 18,428,356 | 18,428,433 | CZRN02000178.1 Marmota marmota marmota genome assembly, contig: chr1_part3, whole genome shotgun sequence | Marmota marmota marmota (Alpine marmot) |
|
|
| 75 | JH393291.1 | 98.50 | full | 14,025,140 | 14,025,217 | JH393291.1 Spermophilus tridecemlineatus unplaced genomic scaffold scaffold00013, whole genome shotgun sequence | Spermophilus tridecemlineatus (thirteen-lined ground squirrel) |
|
|
| 76 | KZ294625.1 | 98.50 | full | 1,304,342 | 1,304,419 | KZ294625.1 Spermophilus dauricus unplaced genomic scaffold scaffold416, whole genome shotgun sequence | Spermophilus dauricus (Daurian ground squirrel) |
|
|
| 77 | QVIC01000060.1 | 98.50 | full | 1,589,376 | 1,589,299 | QVIC01000060.1 Urocitellus parryii isolate AGS 11-09-20 scaffold_59, whole genome shotgun sequence | Spermophilus parryii (Arctic ground squirrel) |
|
|
| 78 | CAJHUB010000666.1 | 98.30 | full | 1,830,041 | 1,829,965 | CAJHUB010000666.1 Nyctereutes procyonoides isolate TBG_1078 genome assembly, contig: Nypro699, whole genome shotgun sequence | Nyctereutes procyonoides (raccoon dog) |
|
|
| 79 | CYRY02030037.1 | 98.10 | full | 2,258 | 2,334 | CYRY02030037.1 Gulo gulo genome assembly, contig: scaffold3867, whole genome shotgun sequence | Gulo gulo (wolverine) |
|
|
| 80 | JAATJU010023977.1 | 97.90 | full | 3,815,481 | 3,815,405 | JAATJU010023977.1 Microtus ochrogaster isolate LTLLF scaffold112, whole genome shotgun sequence | Microtus ochrogaster (prairie vole) |
|
|
| 81 | NW_011907515.1 | 97.30 | full | 720 | 643 | NW_011907515.1 Pteropus vampyrus isolate Shadow unplaced genomic scaffold, Pvam_2.0 Scaffold18734, whole genome shotgun sequence | Pteropus vampyrus (large flying fox) |
|
|
| 82 | JH725464.1 | 96.80 | full | 24,437,617 | 24,437,691 | JH725464.1 Jaculus jaculus unplaced genomic scaffold scaffold00028, whole genome shotgun sequence | Jaculus jaculus (lesser Egyptian jerboa) |
|
|
| 83 | KN122764.1 | 96.60 | full | 623,688 | 623,764 | KN122764.1 Fukomys damarensis unplaced genomic scaffold scaffold318, whole genome shotgun sequence | Fukomys damarensis (Damara mole-rat) |
|
|
| 84 | JH378124.1 | 96.40 | full | 4,765,694 | 4,765,770 | JH378124.1 Saimiri boliviensis boliviensis unplaced genomic scaffold scaffold00020, whole genome shotgun sequence | Saimiri boliviensis boliviensis (Bolivian squirrel monkey) |
|
|
| 85 | JH725480.1 | 96.40 | full | 2,491,641 | 2,491,567 | JH725480.1 Jaculus jaculus unplaced genomic scaffold scaffold00044, whole genome shotgun sequence | Jaculus jaculus (lesser Egyptian jerboa) |
|
|
| 86 | NW_006408618.1 | 96.40 | full | 3,451,366 | 3,451,291 | NW_006408618.1 Chrysochloris asiatica unplaced genomic scaffold, ChrAsi1.0 scaffold00100, whole genome shotgun sequence | Chrysochloris asiatica |
|
|
| 87 | NW_006408618.1 | 96.40 | full | 3,455,132 | 3,455,057 | NW_006408618.1 Chrysochloris asiatica unplaced genomic scaffold, ChrAsi1.0 scaffold00100, whole genome shotgun sequence | Chrysochloris asiatica |
|
|
| 88 | NC_041217.1 | 95.90 | full | 44,403,611 | 44,403,534 | NC_041217.1 Physeter catodon isolate SW-GA chromosome 4, ASM283717v5, whole genome shotgun sequence | Physeter catodon (sperm whale) |
|
|
| 89 | SGJD01003878.1 | 95.90 | full | 211,018 | 210,941 | SGJD01003878.1 Balaenoptera physalus isolate FinWhale-01 scaffold4909, whole genome shotgun sequence | Balaenoptera physalus (Fin whale) |
|
|
| 90 | KB030474.1 | 95.60 | full | 14,220,199 | 14,220,276 | KB030474.1 Pteropus alecto unplaced genomic scaffold scaffold101, whole genome shotgun sequence | Pteropus alecto (black flying fox) |
|
|
| 91 | JH721866.1 | 95.50 | full | 13,995,279 | 13,995,204 | JH721866.1 Chinchilla lanigera unplaced genomic scaffold scaffold00005, whole genome shotgun sequence | Chinchilla lanigera |
|
|
| 92 | KV389598.1 | 95.50 | full | 1,502,650 | 1,502,726 | KV389598.1 Cebus capucinus imitator isolate Cc_AM_T3 unplaced genomic scaffold Scaffold172, whole genome shotgun sequence | Cebus imitator (Panamanian white-faced capuchin) |
|
|
| 93 | NW_022436984.1 | 95.50 | full | 17,805,122 | 17,805,046 | NW_022436984.1 Sapajus apella isolate SASKATOON/1434 unplaced genomic scaffold, GSC_monkey_1.0 scaffold44, whole genome shotgun sequence | Cebus apella (Tufted capuchin) |
|
|
| 94 | NW_007246125.1 | 95.40 | full | 260,717 | 260,793 | NW_007246125.1 Carlito syrichta isolate Samal-C Ts95f unplaced genomic scaffold, Tarsius_syrichta-2.0.1 Scaffold302029, whole genome shotgun sequence | Carlito syrichta (Philippine tarsier) |
|
|
| 95 | QGOO02001649.1 | 94.80 | full | 1,797 | 1,721 | QGOO02001649.1 Mus spicilegus strain ZRU musp714_scaffold_01651, whole genome shotgun sequence | Mus spicilegus (steppe mouse) |
|
|
| 96 | NTJE010278150.1 | 94.80 | full | 16,162 | 16,085 | NTJE010278150.1 Eschrichtius robustus isolate AM-GW-M-EMB-4M-20130531 contig_310808, whole genome shotgun sequence | Eschrichtius robustus (grey whale) |
|
|
| 97 | CM022826.1 | 94.70 | full | 77,585,727 | 77,585,655 | CM022826.1 Ailuropoda melanoleuca isolate Jingjing chromosome 8, whole genome shotgun sequence | Ailuropoda melanoleuca (giant panda) |
|
|
| 98 | JH168899.1 | 94.50 | full | 129,449 | 129,376 | JH168899.1 Heterocephalus glaber unplaced genomic scaffold scaffold2873, whole genome shotgun sequence | Heterocephalus glaber (naked mole-rat) |
|
|
| 99 | MPIZ01000981.1 | 94.20 | full | 1,681,907 | 1,681,828 | MPIZ01000981.1 Prolemur simus strain KIAN8.4 scaffold981, whole genome shotgun sequence | Prolemur simus (greater bamboo lemur) |
|
|
| 100 | LR738623.1 | 93.70 | full | 106,896,384 | 106,896,460 | LR738623.1 Sciurus vulgaris genome assembly, chromosome: 12 | Sciurus vulgaris (Eurasian red squirrel) |
|
|
| 101 | GL010043.1 | 93.50 | full | 27,357,421 | 27,357,494 | Loxodonta africana unplaced genomic scaffold scaffold_16, whole genome shotgun sequence. | Loxodonta africana (African savanna elephant) |
|
|
| 102 | JACASF010000020.1 | 93.40 | full | 48,748,847 | 48,748,923 | JACASF010000020.1 Molossus molossus isolate mMolMol1 scaffold_m16_p_20, whole genome shotgun sequence | Molossus molossus (Pallas's mastiff bat) |
|
|
| 103 | FNWR01000074.1 | 93.20 | full | 403,776 | 403,852 | FNWR01000074.1 Neovison vison genome assembly, contig: scaffold74, whole genome shotgun sequence | Neovison vison |
|
|
| 104 | JAGFMF010011697.1 | 93.10 | full | 1,556,002 | 1,556,078 | JAGFMF010011697.1 Galemys pyrenaicus isolate IBE-C5619 2611377, whole genome shotgun sequence | Galemys pyrenaicus |
|
|
| 105 | JH000079.1 | 93.10 | full | 1,491,161 | 1,491,086 | JH000079.1 Cricetulus griseus cell line CHO-K1 unplaced genomic scaffold scaffold172, whole genome shotgun sequence | Cricetulus griseus (Chinese hamster) |
|
|
| 106 | CM008183.2 | 92.70 | full | 55,234,742 | 55,234,666 | CM008183.2 Bos taurus isolate L1 Dominette 01449 registration number 42190680 breed Hereford chromosome 16, whole genome shotgun sequence | Bos taurus (cattle) |
|
|
| 107 | CM011789.1 | 92.70 | full | 54,609,481 | 54,609,405 | CM011789.1 Bos indicus x Bos taurus breed Angus x Brahman F1 hybrid chromosome 16, whole genome shotgun sequence | Bos indicus x Bos taurus (hybrid cattle) |
|
|
| 108 | NC_032665.1 | 92.70 | full | 52,467,827 | 52,467,751 | NC_032665.1 Bos indicus isolate QUIL7308 breed Nelore chromosome 16, Bos_indicus_1.0, whole genome shotgun sequence | Bos indicus |
|
|
| 109 | CM018482.1 | 92.70 | full | 98,306,392 | 98,306,316 | CM018482.1 Muntiacus reevesi isolate UCam_UCB_Mr chromosome 5, whole genome shotgun sequence | Muntiacus reevesi (Chinese muntjac) |
|
|
| 110 | CM018156.1 | 92.40 | full | 130,434,641 | 130,434,564 | CM018156.1 Phocoena sinus isolate mPhoSin1 chromosome 1, whole genome shotgun sequence | Phocoena sinus (vaquita) |
|
|
| 111 | NC_047034.1 | 92.40 | full | 55,625,255 | 55,625,332 | NC_047034.1 Tursiops truncatus isolate mTurTru1 chromosome 1, mTurTru1.mat.Y, whole genome shotgun sequence | Tursiops truncatus (bottlenosed dolphin) |
|
|
| 112 | NW_020174648.1 | 92.40 | full | 6,636,911 | 6,636,988 | NW_020174648.1 Neophocaena asiaeorientalis asiaeorientalis breed wild unplaced genomic scaffold, Neophocaena_asiaeorientalis_V1 scaffold182, whole genome shotgun sequence | Neophocaena asiaeorientalis asiaeorientalis (yangtze finless porpoise) |
|
|
| 113 | NW_022098095.1 | 92.40 | full | 512,420 | 512,497 | NW_022098095.1 Delphinapterus leucas isolate GAN/ISIS: 26980492/103006 unplaced genomic scaffold, ASM228892v3 scaffoldscaffold104, whole genome shotgun sequence | Delphinapterus leucas (beluga whale) |
|
|
| 114 | JH725438.1 | 92.30 | full | 43,551,076 | 43,551,150 | JH725438.1 Jaculus jaculus unplaced genomic scaffold scaffold00002, whole genome shotgun sequence | Jaculus jaculus (lesser Egyptian jerboa) |
|
|
| 115 | KN977149.1 | 92.30 | full | 5,311,866 | 5,311,789 | KN977149.1 Mandrillus leucophaeus isolate KB7577 unplaced genomic scaffold Scaffold49, whole genome shotgun sequence | Mandrillus leucophaeus (drill) |
|
|
| 116 | NW_012267243.1 | 92.30 | full | 9,871,775 | 9,871,851 | NW_012267243.1 Dipodomys ordii isolate 6190 unplaced genomic scaffold, Dord_2.0 Scaffold27, whole genome shotgun sequence | Dipodomys ordii (Ord's kangaroo rat) |
|
|
| 117 | PVHU020023298.1 | 92.20 | full | 3,825,951 | 3,826,027 | PVHU020023298.1 Moschus moschiferus isolate BS20 Scw3uUT_23202;HRSCAF=125829_4, whole genome shotgun sequence | Moschus moschiferus (Siberian musk deer) |
|
|
| 118 | CM000669.2 | 92.00 | full | 137,287,687 | 137,287,762 | Homo sapiens chromosome 7, GRCh38 reference primary assembly. | Homo sapiens (human) |
|
|
| 119 | CM021205.1 | 92.00 | full | 51,259,182 | 51,259,106 | CM021205.1 Cervus hanglu yarkandensis isolate CEY-2017 chromosome 14, whole genome shotgun sequence | Cervus elaphus yarkandensis |
|
|
| 120 | FR853100.2 | 92.00 | full | 136,749,387 | 136,749,462 | Gorilla gorilla gorilla genomic chromosome, chr7, whole genome shotgun sequence | Gorilla gorilla gorilla (western lowland gorilla) |
|
|
| 121 | KL205491.1 | 92.00 | full | 150,402 | 150,326 | KL205491.1 Nannospalax galili isolate Female #2095 unplaced genomic scaffold scaffold429, whole genome shotgun sequence | Nannospalax galili (Upper Galilee mountains blind mole rat) |
|
|
| 122 | CM001593.2 | 92.00 | full | 53,200,656 | 53,200,580 | Ovis aries breed Texel chromosome 12, whole genome shotgun sequence. | Ovis aries (sheep) |
|
|
| 123 | CM004577.1 | 92.00 | full | 53,756,365 | 53,756,289 | CM004577.1 Capra hircus breed San Clemente chromosome 16, whole genome shotgun sequence | Capra hircus (goat) |
|
|
| 124 | CM008021.1 | 92.00 | full | 70,395,592 | 70,395,516 | CM008021.1 Cervus elaphus hippelaphus isolate Hungarian chromosome 14, whole genome shotgun sequence | Cervus elaphus hippelaphus |
|
|
| 125 | CM000996.3 | 91.90 | full | 100,210,604 | 100,210,680 | CM000996.3 Mus musculus chromosome 3, GRCm39 reference primary assembly C57BL/6J | Mus musculus (house mouse) |
|
|
| 126 | NW_006799328.1 | 91.80 | full | 212,473 | 212,396 | NW_006799328.1 Lipotes vexillifer unplaced genomic scaffold, Lipotes_vexillifer_v1 scaffold821, whole genome shotgun sequence | Lipotes vexillifer (Yangtze River dolphin) |
|
|
| 127 | CM009269.2 | 91.60 | full | 123,287,088 | 123,287,163 | CM009269.2 Pongo abelii isolate Susie chromosome 7, whole genome shotgun sequence | Pongo abelii (Sumatran orangutan) |
|
|
| 128 | NC_045785.1 | 91.40 | full | 129,459,314 | 129,459,237 | NC_045785.1 Balaenoptera musculus isolate JJ_BM4_2016_0621 chromosome 1, mBalMus1.pri.v3, whole genome shotgun sequence | Balaenoptera musculus (Blue whale) |
|
|
| 129 | CM009152.1 | 91.30 | full | 9,553,495 | 9,553,419 | CM009152.1 Equus caballus isolate Twilight breed thoroughbred chromosome 5, whole genome shotgun sequence | Equus caballus (horse) |
|
|
| 130 | CM027714.2 | 91.30 | full | 37,589,487 | 37,589,563 | CM027714.2 Equus asinus breed Dezhou chromosome 25, whole genome shotgun sequence | Equus asinus (ass) |
|
|
| 131 | CM001658.1 | 91.10 | full | 90,941,643 | 90,941,567 | Nomascus leucogenys chromosome 12, whole genome shotgun sequence. | Nomascus leucogenys (Northern white-cheeked gibbon) |
|
|
| 132 | NW_004524593.1 | 90.90 | full | 3,609,764 | 3,609,838 | NW_004524593.1 Octodon degus isolate 3935 unplaced genomic scaffold, OctDeg1.0 scaffold00017, whole genome shotgun sequence | Octodon degus (degu) |
|
|
| 133 | NC_040916.2 | 90.90 | full | 21,824,145 | 21,824,220 | NC_040916.2 Phyllostomus discolor isolate MPI-MPIP mPhyDis1 chromosome 14, mPhyDis1.pri.v3, whole genome shotgun sequence | Phyllostomus discolor (pale spear-nosed bat) |
|
|
| 134 | QWLN02015690.1 | 90.70 | full | 1,082,371 | 1,082,294 | QWLN02015690.1 Sousa chinensis isolate MY-2018 scaffold181.1, whole genome shotgun sequence | Sousa chinensis (Indo-pacific humpbacked dolphin) |
|
|
| 135 | CM009262.2 | 90.60 | full | 141,673,797 | 141,673,721 | CM009262.2 Pongo abelii isolate Susie chromosome 1, whole genome shotgun sequence | Pongo abelii (Sumatran orangutan) |
|
|
| 136 | JH725458.1 | 90.50 | full | 10,845,687 | 10,845,613 | JH725458.1 Jaculus jaculus unplaced genomic scaffold scaffold00022, whole genome shotgun sequence | Jaculus jaculus (lesser Egyptian jerboa) |
|
|
| 137 | CM000321.4 | 90.00 | full | 141,871,154 | 141,871,229 | Pan troglodytes isolate Yerkes chimp pedigree #C0471 (Clint) chromosome 7, whole genome shotgun sequence. | Pan troglodytes (chimpanzee) |
|
|
| 138 | CM003390.1 | 90.00 | full | 141,737,955 | 141,738,030 | CM003390.1 Pan paniscus isolate Ulindi chromosome 7, whole genome shotgun sequence | Pan paniscus (pygmy chimpanzee) |
|
|
| 139 | RWIC01001038.1 | 89.90 | full | 264,604 | 264,527 | RWIC01001038.1 Monodon monoceros isolate MVW Scaffold1038, whole genome shotgun sequence | Monodon monoceros (narwhal) |
|
|
| 140 | CM000820.5 | 89.80 | full | 116,145,390 | 116,145,312 | Sus scrofa isolate TJ Tabasco breed Duroc chromosome 9, whole genome shotgun sequence. | Sus scrofa (pig) |
|
|
| 141 | NW_018335398.1 | 89.60 | full | 819,023 | 818,947 | NW_018335398.1 Odocoileus virginianus texanus isolate animal Pink-7 unplaced genomic scaffold, Ovir.te_1.0 scaffold23, whole genome shotgun sequence | Odocoileus virginianus texanus |
|
|
| 142 | JH168479.1 | 89.40 | full | 1,376,207 | 1,376,131 | JH168479.1 Heterocephalus glaber unplaced genomic scaffold scaffold823, whole genome shotgun sequence | Heterocephalus glaber (naked mole-rat) |
|
|
| 143 | CM014341.1 | 88.60 | full | 18,326,272 | 18,326,348 | CM014341.1 Macaca mulatta isolate AG07107 chromosome 6, whole genome shotgun sequence | Macaca mulatta (Rhesus monkey) |
|
|
| 144 | CM016703.1 | 88.60 | full | 53,846,832 | 53,846,756 | CM016703.1 Bos grunniens isolate yakHY00 ecotype maiwa chromosome 14, whole genome shotgun sequence | Bos grunniens (domestic yak) |
|
|
| 145 | CM021944.1 | 88.60 | full | 18,398,407 | 18,398,483 | CM021944.1 Macaca fascicularis isolate cy0333 breed Mauritian cynomolgus macaque chromosome 6, whole genome shotgun sequence | Macaca fascicularis (crab-eating macaque) |
|
|
| 146 | JH880647.1 | 88.60 | full | 1,318,943 | 1,319,019 | JH880647.1 Bos grunniens mutus unplaced genomic scaffold scaffold341_1, whole genome shotgun sequence | Bos grunniens mutus |
|
|
| 147 | KQ005458.1 | 88.60 | full | 1,272,435 | 1,272,359 | KQ005458.1 Macaca nemestrina isolate M95218 unplaced genomic scaffold Scaffold178, whole genome shotgun sequence | Macaca nemestrina (pig-tailed macaque) |
|
|
| 148 | NW_011494861.1 | 88.60 | full | 15,391,372 | 15,391,296 | NW_011494861.1 Bison bison bison isolate TAMUID 2011002044 unplaced genomic scaffold, Bison_UMD1.0 scf7180017864289, whole genome shotgun sequence | Bison bison bison (north american plains bison) |
|
|
| 149 | NW_006921762.1 | 88.00 | full | 3,583,271 | 3,583,344 | NW_006921762.1 Orycteropus afer afer isolate SDZICR_OR568_19922 unplaced genomic scaffold, OryAfe1.0 scaffold00175, whole genome shotgun sequence | Orycteropus afer afer |
|
|
| 150 | NW_004443957.1 | 87.70 | full | 10,410,307 | 10,410,231 | NW_004443957.1 Trichechus manatus latirostris isolate Lorelei unplaced genomic scaffold, TriManLat1.0 scaffold00021, whole genome shotgun sequence | Trichechus manatus latirostris |
|
|
| 151 | DS562867.1 | 87.60 | full | 10,039,941 | 10,040,020 | Cavia porcellus supercont2_12 genomic scaffold, whole genome shotgun sequence. | Cavia porcellus (Domestic guinea pig) |
|
|
| 152 | CM009952.1 | 87.50 | full | 161,798,877 | 161,798,954 | CM009952.1 Theropithecus gelada isolate Dixy chromosome 3, whole genome shotgun sequence | Theropithecus gelada (gelada baboon) |
|
|
| 153 | CM014338.1 | 87.50 | full | 163,708,559 | 163,708,636 | CM014338.1 Macaca mulatta isolate AG07107 chromosome 3, whole genome shotgun sequence | Macaca mulatta (Rhesus monkey) |
|
|
| 154 | CM018183.2 | 87.50 | full | 21,358,865 | 21,358,788 | CM018183.2 Papio anubis isolate 15944 chromosome 4, whole genome shotgun sequence | Papio anubis (Olive baboon) |
|
|
| 155 | CM021941.1 | 87.50 | full | 164,722,862 | 164,722,939 | CM021941.1 Macaca fascicularis isolate cy0333 breed Mauritian cynomolgus macaque chromosome 3, whole genome shotgun sequence | Macaca fascicularis (crab-eating macaque) |
|
|
| 156 | KN299787.1 | 87.50 | full | 3,212,625 | 3,212,548 | KN299787.1 Rhinopithecus roxellana isolate Xiao Hai unplaced genomic scaffold ENSRROG047212, whole genome shotgun sequence | Rhinopithecus roxellana (golden snub-nosed monkey) |
|
|
| 157 | KN980495.1 | 87.50 | full | 5,353,184 | 5,353,261 | KN980495.1 Colobus angolensis palliatus isolate OR3802 unplaced genomic scaffold Scaffold2, whole genome shotgun sequence | Colobus angolensis palliatus |
|
|
| 158 | KQ008339.1 | 87.50 | full | 7,067,184 | 7,067,107 | KQ008339.1 Macaca nemestrina isolate M95218 unplaced genomic scaffold Scaffold46, whole genome shotgun sequence | Macaca nemestrina (pig-tailed macaque) |
|
|
| 159 | KQ009908.1 | 87.50 | full | 2,827,786 | 2,827,863 | KQ009908.1 Cercocebus atys isolate FAK unplaced genomic scaffold Scaffold137, whole genome shotgun sequence | Cercocebus atys (sooty mangabey) |
|
|
| 160 | MCGX01003620.1 | 87.50 | full | 2,411,072 | 2,411,149 | MCGX01003620.1 Rhinopithecus bieti isolate Rb0 scaffold_8307, whole genome shotgun sequence | Rhinopithecus bieti (Black snub-nosed monkey) |
|
|
| 161 | PVHT020000175.1 | 87.50 | full | 8,417,161 | 8,417,085 | PVHT020000175.1 Catagonus wagneri isolate BS18 Sc2sB16_147;HRSCAF=32195_2, whole genome shotgun sequence | Catagonus wagneri (Chacoan peccary) |
|
|
| 162 | KN981225.1 | 87.30 | full | 1,666,816 | 1,666,740 | KN981225.1 Colobus angolensis palliatus isolate OR3802 unplaced genomic scaffold Scaffold266, whole genome shotgun sequence | Colobus angolensis palliatus |
|
|
| 163 | CM021922.1 | 86.70 | full | 107,307,285 | 107,307,210 | CM021922.1 Callithrix jacchus isolate mCalJac1 chromosome 8, whole genome shotgun sequence | Callithrix jacchus (white-tufted-ear marmoset) |
|
|
| 164 | CM000663.2 | 86.10 | full | 85,592,280 | 85,592,356 | Homo sapiens chromosome 1, GRCh38 reference primary assembly. | Homo sapiens (human) |
|
|
| 165 | FR853080.2 | 86.10 | full | 86,514,860 | 86,514,936 | Gorilla gorilla gorilla genomic chromosome, chr1, whole genome shotgun sequence | Gorilla gorilla gorilla (western lowland gorilla) |
|
|
| 166 | CM019243.1 | 85.70 | full | 17,630,667 | 17,630,743 | CM019243.1 Piliocolobus tephrosceles isolate RC106 chromosome 4, whole genome shotgun sequence | Piliocolobus tephrosceles (Ugandan red Colobus) |
|
|
| 167 | CM018184.2 | 85.60 | full | 17,421,262 | 17,421,338 | CM018184.2 Papio anubis isolate 15944 chromosome 5, whole genome shotgun sequence | Papio anubis (Olive baboon) |
|
|
| 168 | KQ010425.1 | 85.60 | full | 589,196 | 589,272 | KQ010425.1 Cercocebus atys isolate FAK unplaced genomic scaffold Scaffold186, whole genome shotgun sequence | Cercocebus atys (sooty mangabey) |
|
|
| 169 | NW_006804477.1 | 85.60 | full | 106,187 | 106,261 | NW_006804477.1 Erinaceus europaeus isolate Erinaceus europaeus_13Jul2011 unplaced genomic scaffold, EriEur2.0 scaffold00554, whole genome shotgun sequence | Erinaceus europaeus (western European hedgehog) |
|
|
| 170 | JH378123.1 | 85.40 | full | 8,615,036 | 8,615,112 | JH378123.1 Saimiri boliviensis boliviensis unplaced genomic scaffold scaffold00019, whole genome shotgun sequence | Saimiri boliviensis boliviensis (Bolivian squirrel monkey) |
|
|
| 171 | CM001659.1 | 85.20 | full | 88,566,614 | 88,566,689 | Nomascus leucogenys chromosome 13, whole genome shotgun sequence. | Nomascus leucogenys (Northern white-cheeked gibbon) |
|
|
| 172 | CM019247.1 | 85.20 | full | 126,411,081 | 126,411,158 | CM019247.1 Piliocolobus tephrosceles isolate RC106 chromosome 8, whole genome shotgun sequence | Piliocolobus tephrosceles (Ugandan red Colobus) |
|
|
| 173 | VOAJ01023713.1 | 84.80 | full | 1,124 | 1,050 | VOAJ01023713.1 Crocuta crocuta isolate KB4526 C28859344, whole genome shotgun sequence | Crocuta crocuta (spotted hyena) |
|
|
| 174 | KQ021358.1 | 84.70 | full | 2,076,409 | 2,076,485 | KQ021358.1 Propithecus coquereli isolate 6110/MARCELLA unplaced genomic scaffold Scaffold186, whole genome shotgun sequence | Propithecus coquereli (Coquerel's sifaka) |
|
|
| 175 | JH378140.1 | 84.50 | full | 3,596,143 | 3,596,218 | JH378140.1 Saimiri boliviensis boliviensis unplaced genomic scaffold scaffold00036, whole genome shotgun sequence | Saimiri boliviensis boliviensis (Bolivian squirrel monkey) |
|
|
| 176 | KN299790.1 | 84.40 | full | 1,830,424 | 1,830,500 | KN299790.1 Rhinopithecus roxellana isolate Xiao Hai unplaced genomic scaffold ENSRROG047218, whole genome shotgun sequence | Rhinopithecus roxellana (golden snub-nosed monkey) |
|
|
| 177 | MCGX01018405.1 | 84.40 | full | 1,330,295 | 1,330,371 | MCGX01018405.1 Rhinopithecus bieti isolate Rb0 scaffold_52030, whole genome shotgun sequence | Rhinopithecus bieti (Black snub-nosed monkey) |
|
|
| 178 | DS562893.1 | 84.20 | full | 21,197,841 | 21,197,917 | Cavia porcellus supercont2_38 genomic scaffold, whole genome shotgun sequence. | Cavia porcellus (Domestic guinea pig) |
|
|
| 179 | CM000314.3 | 83.70 | full | 86,019,912 | 86,019,988 | Pan troglodytes isolate Yerkes chimp pedigree #C0471 (Clint) chromosome 1, whole genome shotgun sequence. | Pan troglodytes (chimpanzee) |
|
|
| 180 | CM003383.1 | 83.70 | full | 87,005,689 | 87,005,765 | CM003383.1 Pan paniscus isolate Ulindi chromosome 1, whole genome shotgun sequence | Pan paniscus (pygmy chimpanzee) |
|
|
| 181 | CM001962.2 | 83.30 | full | 105,970,048 | 105,970,125 | Chlorocebus sabaeus isolate 1994-021 chromosome 21, whole genome shotgun sequence. | Chlorocebus sabaeus |
|
|
| 182 | JH725437.1 | 83.10 | full | 19,614,127 | 19,614,201 | JH725437.1 Jaculus jaculus unplaced genomic scaffold scaffold00001, whole genome shotgun sequence | Jaculus jaculus (lesser Egyptian jerboa) |
|
|
| 183 | JH725437.1 | 82.60 | full | 69,745,717 | 69,745,792 | JH725437.1 Jaculus jaculus unplaced genomic scaffold scaffold00001, whole genome shotgun sequence | Jaculus jaculus (lesser Egyptian jerboa) |
|
|
| 184 | NW_004524643.1 | 82.50 | full | 12,141,277 | 12,141,203 | NW_004524643.1 Octodon degus isolate 3935 unplaced genomic scaffold, OctDeg1.0 scaffold00067, whole genome shotgun sequence | Octodon degus (degu) |
|
|
| 185 | KN974960.1 | 82.40 | full | 612,784 | 612,860 | KN974960.1 Mandrillus leucophaeus isolate KB7577 unplaced genomic scaffold Scaffold278, whole genome shotgun sequence | Mandrillus leucophaeus (drill) |
|
|
| 186 | CM019251.1 | 81.80 | full | 11,838,023 | 11,838,099 | CM019251.1 Piliocolobus tephrosceles isolate RC106 chromosome 12, whole genome shotgun sequence | Piliocolobus tephrosceles (Ugandan red Colobus) |
|
|
| 187 | NW_012267234.1 | 81.70 | full | 4,277,804 | 4,277,728 | NW_012267234.1 Dipodomys ordii isolate 6190 unplaced genomic scaffold, Dord_2.0 Scaffold18, whole genome shotgun sequence | Dipodomys ordii (Ord's kangaroo rat) |
|
|
| 188 | CM009971.1 | 81.60 | full | 138,382,340 | 138,382,416 | CM009971.1 Theropithecus gelada isolate Dixy chromosome X, whole genome shotgun sequence | Theropithecus gelada (gelada baboon) |
|
|
| 189 | CM014356.1 | 81.60 | full | 137,018,820 | 137,018,896 | CM014356.1 Macaca mulatta isolate AG07107 chromosome X, whole genome shotgun sequence | Macaca mulatta (Rhesus monkey) |
|
|
| 190 | CM018200.1 | 81.60 | full | 13,786,723 | 13,786,647 | CM018200.1 Papio anubis isolate 15944 chromosome X, whole genome shotgun sequence | Papio anubis (Olive baboon) |
|
|
| 191 | CM021959.1 | 81.60 | full | 134,067,503 | 134,067,579 | CM021959.1 Macaca fascicularis isolate cy0333 breed Mauritian cynomolgus macaque chromosome X, whole genome shotgun sequence | Macaca fascicularis (crab-eating macaque) |
|
|
| 192 | KN979181.1 | 81.60 | full | 911,979 | 911,903 | KN979181.1 Mandrillus leucophaeus isolate KB7577 unplaced genomic scaffold Scaffold816, whole genome shotgun sequence | Mandrillus leucophaeus (drill) |
|
|
| 193 | KQ006312.1 | 81.60 | full | 1,141,896 | 1,141,820 | KQ006312.1 Macaca nemestrina isolate M95218 unplaced genomic scaffold Scaffold257, whole genome shotgun sequence | Macaca nemestrina (pig-tailed macaque) |
|
|
| 194 | KQ010259.1 | 81.60 | full | 5,719,219 | 5,719,295 | KQ010259.1 Cercocebus atys isolate FAK unplaced genomic scaffold Scaffold170, whole genome shotgun sequence | Cercocebus atys (sooty mangabey) |
|
|
| 195 | CM009955.1 | 81.40 | full | 18,101,433 | 18,101,509 | CM009955.1 Theropithecus gelada isolate Dixy chromosome 6, whole genome shotgun sequence | Theropithecus gelada (gelada baboon) |
|
|
| 196 | CM001652.1 | 81.30 | full | 17,236,456 | 17,236,380 | Nomascus leucogenys chromosome 6, whole genome shotgun sequence. | Nomascus leucogenys (Northern white-cheeked gibbon) |
|
|
| 197 | MPIZ01001698.1 | 80.30 | full | 1,967,529 | 1,967,453 | MPIZ01001698.1 Prolemur simus strain KIAN8.4 scaffold1698, whole genome shotgun sequence | Prolemur simus (greater bamboo lemur) |
|
|
| 198 | CM009267.2 | 79.70 | full | 18,562,817 | 18,562,893 | CM009267.2 Pongo abelii isolate Susie chromosome 5, whole genome shotgun sequence | Pongo abelii (Sumatran orangutan) |
|
|
| 199 | NW_004801655.1 | 79.20 | full | 10,138,765 | 10,138,697 | NW_004801655.1 Mesocricetus auratus isolate Golden Hamster female 1 unplaced genomic scaffold, MesAur1.0 scaffold00052, whole genome shotgun sequence | Mesocricetus auratus (golden hamster) |
|
|
| 200 | KZ196488.1 | 79.10 | full | 2,773,697 | 2,773,772 | KZ196488.1 Aotus nancymaae isolate 86115 unplaced genomic scaffold Scaffold114.225, whole genome shotgun sequence | Aotus nancymaae (Ma's night monkey) |
|
|
| 201 | NW_004525003.1 | 78.20 | full | 132,113 | 132,038 | NW_004525003.1 Octodon degus isolate 3935 unplaced genomic scaffold, OctDeg1.0 scaffold00427, whole genome shotgun sequence | Octodon degus (degu) |
|
|
| 202 | KV389745.1 | 77.20 | full | 1,478,728 | 1,478,653 | KV389745.1 Cebus capucinus imitator isolate Cc_AM_T3 unplaced genomic scaffold Scaffold319, whole genome shotgun sequence | Cebus imitator (Panamanian white-faced capuchin) |
|
|
| 203 | NW_004525003.1 | 76.90 | full | 56,035 | 55,961 | NW_004525003.1 Octodon degus isolate 3935 unplaced genomic scaffold, OctDeg1.0 scaffold00427, whole genome shotgun sequence | Octodon degus (degu) |
|
|
| 204 | CM001944.2 | 76.00 | full | 17,721,493 | 17,721,569 | Chlorocebus sabaeus isolate 1994-021 chromosome 4, whole genome shotgun sequence. | Chlorocebus sabaeus |
|
|
| 205 | MPIZ01000292.1 | 75.90 | full | 229,349 | 229,423 | MPIZ01000292.1 Prolemur simus strain KIAN8.4 scaffold292, whole genome shotgun sequence | Prolemur simus (greater bamboo lemur) |
|
|
| 206 | CM007671.1 | 75.80 | full | 15,330,798 | 15,330,722 | CM007671.1 Microcebus murinus isolate mixed chromosome 11, whole genome shotgun sequence | Microcebus murinus (gray mouse lemur) |
|
|
| 207 | CM007673.1 | 74.70 | full | 46,422,778 | 46,422,704 | CM007673.1 Microcebus murinus isolate mixed chromosome 13, whole genome shotgun sequence | Microcebus murinus (gray mouse lemur) |
|
|
| 208 | KN298748.1 | 74.40 | full | 301,425 | 301,349 | KN298748.1 Rhinopithecus roxellana isolate Xiao Hai unplaced genomic scaffold ENSRROG043834, whole genome shotgun sequence | Rhinopithecus roxellana (golden snub-nosed monkey) |
|
|
| 209 | MCGX01004714.1 | 73.90 | full | 126,864 | 126,788 | MCGX01004714.1 Rhinopithecus bieti isolate Rb0 scaffold_13749, whole genome shotgun sequence | Rhinopithecus bieti (Black snub-nosed monkey) |
|
|
| 210 | NW_022436951.1 | 73.70 | full | 23,902,847 | 23,902,771 | NW_022436951.1 Sapajus apella isolate SASKATOON/1434 unplaced genomic scaffold, GSC_monkey_1.0 scaffold11, whole genome shotgun sequence | Cebus apella (Tufted capuchin) |
|
|
| 211 | KN980274.1 | 73.30 | full | 5,735,501 | 5,735,577 | KN980274.1 Colobus angolensis palliatus isolate OR3802 unplaced genomic scaffold Scaffold180, whole genome shotgun sequence | Colobus angolensis palliatus |
|
|
| 212 | CM003388.1 | 73.20 | full | 18,513,024 | 18,513,097 | CM003388.1 Pan paniscus isolate Ulindi chromosome 5, whole genome shotgun sequence | Pan paniscus (pygmy chimpanzee) |
|
|
| 213 | FR853088.3 | 73.20 | full | 77,339,650 | 77,339,577 | Gorilla gorilla gorilla genomic chromosome, chr17, whole genome shotgun sequence | Gorilla gorilla gorilla (western lowland gorilla) |
|
|
| 214 | JH721943.1 | 72.60 | full | 327,516 | 327,440 | JH721943.1 Chinchilla lanigera unplaced genomic scaffold scaffold00082, whole genome shotgun sequence | Chinchilla lanigera |
|
|
| 215 | JH721866.1 | 72.40 | full | 8,982,292 | 8,982,367 | JH721866.1 Chinchilla lanigera unplaced genomic scaffold scaffold00005, whole genome shotgun sequence | Chinchilla lanigera |
|
|
| 216 | NW_004525048.1 | 72.30 | full | 82,931 | 82,857 | NW_004525048.1 Octodon degus isolate 3935 unplaced genomic scaffold, OctDeg1.0 scaffold00472, whole genome shotgun sequence | Octodon degus (degu) |
|
|
| 217 | KV389461.1 | 72.00 | full | 5,986,113 | 5,986,037 | KV389461.1 Cebus capucinus imitator isolate Cc_AM_T3 unplaced genomic scaffold Scaffold35, whole genome shotgun sequence | Cebus imitator (Panamanian white-faced capuchin) |
|
|
| 218 | CM000319.3 | 71.60 | full | 18,519,674 | 18,519,747 | Pan troglodytes isolate Yerkes chimp pedigree #C0471 (Clint) chromosome 5, whole genome shotgun sequence. | Pan troglodytes (chimpanzee) |
|
|
| 219 | NW_022436947.1 | 70.60 | full | 4,576,942 | 4,576,868 | NW_022436947.1 Sapajus apella isolate SASKATOON/1434 unplaced genomic scaffold, GSC_monkey_1.0 scaffold7, whole genome shotgun sequence | Cebus apella (Tufted capuchin) |
|
|
| 220 | JH166615.1 | 70.00 | full | 315,350 | 315,273 | JH166615.1 Heterocephalus glaber unplaced genomic scaffold scaffold1481, whole genome shotgun sequence | Heterocephalus glaber (naked mole-rat) |
|
|
| 221 | JH166615.1 | 70.00 | full | 316,641 | 316,564 | JH166615.1 Heterocephalus glaber unplaced genomic scaffold scaffold1481, whole genome shotgun sequence | Heterocephalus glaber (naked mole-rat) |
|
|
| 222 | KZ202417.1 | 69.90 | full | 18,828,215 | 18,828,291 | KZ202417.1 Aotus nancymaae isolate 86115 unplaced genomic scaffold Scaffold31, whole genome shotgun sequence | Aotus nancymaae (Ma's night monkey) |
|
|
| 223 | NW_007251453.1 | 69.50 | full | 6,799 | 6,723 | NW_007251453.1 Carlito syrichta isolate Samal-C Ts95f unplaced genomic scaffold, Tarsius_syrichta-2.0.1 Scaffold307357, whole genome shotgun sequence | Carlito syrichta (Philippine tarsier) |
|
|
| 224 | CM000667.2 | 69.20 | full | 18,236,123 | 18,236,196 | Homo sapiens chromosome 5, GRCh38 reference primary assembly. | Homo sapiens (human) |
|
|
| 225 | JH378273.1 | 67.70 | full | 1,885,295 | 1,885,371 | JH378273.1 Saimiri boliviensis boliviensis unplaced genomic scaffold scaffold00169, whole genome shotgun sequence | Saimiri boliviensis boliviensis (Bolivian squirrel monkey) |
|
|
| 226 | KB360289.1 | 67.00 | full | 304,011 | 303,935 | KB360289.1 Tupaia chinensis unplaced genomic scaffold Scaffold139457_1, whole genome shotgun sequence | Tupaia chinensis (Chinese tree shrew) |
|
|
| 227 | CM001951.2 | 66.40 | full | 115,932,385 | 115,932,456 | Chlorocebus sabaeus isolate 1994-021 chromosome X, whole genome shotgun sequence. | Chlorocebus sabaeus |
|
|
| 228 | CM021916.1 | 64.90 | full | 183,795,209 | 183,795,133 | CM021916.1 Callithrix jacchus isolate mCalJac1 chromosome 2, whole genome shotgun sequence | Callithrix jacchus (white-tufted-ear marmoset) |
|
|
| 229 | JH174161.1 | 64.10 | full | 629,070 | 628,997 | JH174161.1 Heterocephalus glaber unplaced genomic scaffold scaffold99, whole genome shotgun sequence | Heterocephalus glaber (naked mole-rat) |
|
|
| 230 | JH725453.1 | 63.70 | full | 25,821,395 | 25,821,323 | JH725453.1 Jaculus jaculus unplaced genomic scaffold scaffold00017, whole genome shotgun sequence | Jaculus jaculus (lesser Egyptian jerboa) |
|
|
| 231 | CM025136.1 | 62.60 | full | 12,309,349 | 12,309,439 | CM025136.1 Canis lupus familiaris isolate SID07034 breed Labrador retriever chromosome 37, whole genome shotgun sequence | Canis lupus familiaris (dog) |
|
|
| 232 | CM025493.1 | 62.60 | full | 12,717,056 | 12,717,146 | CM025493.1 Canis lupus dingo isolate Sandy chromosome 37, whole genome shotgun sequence | Canis lupus dingo (dingo) |
|
|
| 233 | JAGFMF010011614.1 | 62.60 | full | 11,358,475 | 11,358,398 | JAGFMF010011614.1 Galemys pyrenaicus isolate IBE-C5619 2611294, whole genome shotgun sequence | Galemys pyrenaicus |
|
|
| 234 | NW_007245732.1 | 59.70 | full | 814,172 | 814,097 | NW_007245732.1 Carlito syrichta isolate Samal-C Ts95f unplaced genomic scaffold, Tarsius_syrichta-2.0.1 Scaffold301636, whole genome shotgun sequence | Carlito syrichta (Philippine tarsier) |
|
|
| 235 | KN122054.1 | 59.50 | full | 22,112,252 | 22,112,326 | KN122054.1 Fukomys damarensis unplaced genomic scaffold scaffold185, whole genome shotgun sequence | Fukomys damarensis (Damara mole-rat) |
|
|
| 236 | JAPFRF010000005.1 | 59.30 | full | 4,082,610 | 4,082,534 | JAPFRF010000005.1 Phrynocephalus forsythii HIC_ASM_4, whole genome shotgun sequence | Phrynocephalus forsythii |
|
|
| 237 | JH725477.1 | 59.20 | full | 8,558,669 | 8,558,599 | JH725477.1 Jaculus jaculus unplaced genomic scaffold scaffold00041, whole genome shotgun sequence | Jaculus jaculus (lesser Egyptian jerboa) |
|
|
| 238 | QVOM02000034.1 | 56.80 | full | 132,113,348 | 132,113,273 | QVOM02000034.1 Salvator merianae strain LG2117 Scaffold_34, whole genome shotgun sequence | Tupinambis merianae |
|
|
| 239 | CM014748.1 | 56.60 | full | 2,848,903 | 2,848,824 | CM014748.1 Podarcis muralis ecotype yellow chromosome 6, whole genome shotgun sequence | Podarcis muralis |
|
|
| 240 | CM014748.1 | 56.60 | full | 2,881,645 | 2,881,566 | CM014748.1 Podarcis muralis ecotype yellow chromosome 6, whole genome shotgun sequence | Podarcis muralis |
|
|
| 241 | OX395131.1 | 56.60 | full | 3,067,411 | 3,067,332 | OX395131.1 Podarcis lilfordi genome assembly, chromosome: 6 | Podarcis lilfordi |
|
|
| 242 | NW_004524695.1 | 56.40 | full | 362,203 | 362,128 | NW_004524695.1 Octodon degus isolate 3935 unplaced genomic scaffold, OctDeg1.0 scaffold00119, whole genome shotgun sequence | Octodon degus (degu) |
|
|
| 243 | JH721906.1 | 56.20 | full | 9,611,641 | 9,611,565 | JH721906.1 Chinchilla lanigera unplaced genomic scaffold scaffold00045, whole genome shotgun sequence | Chinchilla lanigera |
|
|
| 244 | GL873568.1 | 55.70 | full | 7,347,151 | 7,347,076 | Otolemur garnettii unplaced genomic scaffold scaffold00049, whole genome shotgun sequence. | Otolemur garnettii (small-eared galago) |
|
|
| 245 | VZTY01011308.1 | 55.30 | full | 43,244 | 43,319 | VZTY01011308.1 Turnix velox voucher MVMA:130 isolate B10K-DU-029-46 scaffold26153, whole genome shotgun sequence | Turnix velox (little buttonquail) |
|
|
| 246 | NW_006408570.1 | 55.10 | full | 12,822,510 | 12,822,580 | NW_006408570.1 Chrysochloris asiatica unplaced genomic scaffold, ChrAsi1.0 scaffold00052, whole genome shotgun sequence | Chrysochloris asiatica |
|
|
| 247 | VXBH01008625.1 | 55.10 | full | 52,554 | 52,629 | VXBH01008625.1 Rynchops niger voucher USNM:637251 isolate B10K-DU-002-16 scaffold674, whole genome shotgun sequence | Rynchops niger (black skimmer) |
|
|
| 248 | VXBO01010082.1 | 54.70 | full | 16,000 | 15,924 | VXBO01010082.1 Rhinoptilus africanus voucher USNM:642451 isolate B10K-DU-002-36 scaffold5225, whole genome shotgun sequence | Rhinoptilus africanus (double-banded courser) |
|
|
| 249 | CM000940.1 | 54.50 | full | 71,380,400 | 71,380,323 | Anolis carolinensis chromosome 4, whole genome shotgun sequence. | Anolis carolinensis (green anole) |
|
|
| 250 | VYZM01004398.1 | 54.30 | full | 3,086 | 3,161 | VYZM01004398.1 Dromas ardeola voucher USNM:647762 isolate B10K-DU-012-55 scaffold3011, whole genome shotgun sequence | Dromas ardeola |
|
|
| 251 | VZZW01000271.1 | 54.30 | full | 891,603 | 891,528 | VZZW01000271.1 Phaetusa simplex voucher USNM:637171 isolate B10K-DU-009-16 scaffold_244, whole genome shotgun sequence | Phaetusa simplex (large-billed tern) |
|
|
| 252 | NW_018150936.1 | 54.20 | full | 950,574 | 950,497 | NW_018150936.1 Pogona vitticeps unplaced genomic scaffold, pvi1.1.0 contig30912, whole genome shotgun sequence | Pogona vitticeps (central bearded dragon) |
|
|
| 253 | KK841893.1 | 54.10 | full | 5,930 | 6,003 | KK841893.1 Acanthisitta chloris isolate BGI_N310 unplaced genomic scaffold scaffold29237, whole genome shotgun sequence | Acanthisitta chloris |
|
|
| 254 | VWPW01005245.1 | 54.00 | full | 172,958 | 172,880 | VWPW01005245.1 Crypturellus undulatus voucher MSB:37135 isolate B10K-MSB-37135 scaffold36, whole genome shotgun sequence | Crypturellus undulatus |
|
|
| 255 | CM009951.1 | 53.90 | full | 150,016,347 | 150,016,411 | CM009951.1 Theropithecus gelada isolate Dixy chromosome 2, whole genome shotgun sequence | Theropithecus gelada (gelada baboon) |
|
|
| 256 | CM018181.1 | 53.90 | full | 149,661,520 | 149,661,584 | CM018181.1 Papio anubis isolate 15944 chromosome 2, whole genome shotgun sequence | Papio anubis (Olive baboon) |
|
|
| 257 | KN974384.1 | 53.70 | full | 183,796 | 183,749 | KN974384.1 Mandrillus leucophaeus isolate KB7577 unplaced genomic scaffold Scaffold226, whole genome shotgun sequence | Mandrillus leucophaeus (drill) |
|
|
| 258 | KV943259.1 | 53.60 | full | 46,688 | 46,617 | KV943259.1 Rana catesbeiana isolate Bruno unplaced genomic scaffold Rc-01r160223s0021991, whole genome shotgun sequence | Rana catesbeiana (bullfrog) |
|
|
| 259 | JH224678.1 | 53.50 | full | 3,754,762 | 3,754,840 | Pelodiscus sinensis unplaced genomic scaffold scaffold281, whole genome shotgun sequence. | Pelodiscus sinensis (Chinese softshell turtle) |
|
|
| 260 | JH170974.1 | 53.20 | full | 173,159 | 173,084 | JH170974.1 Heterocephalus glaber unplaced genomic scaffold scaffold16, whole genome shotgun sequence | Heterocephalus glaber (naked mole-rat) |
|
|
| 261 | VZSD01013135.1 | 53.00 | full | 151,742 | 151,667 | VZSD01013135.1 Alca torda voucher C:Vinge78 isolate OUT-0003 scaffold4998, whole genome shotgun sequence | Alca torda |
|
|
| 262 | VXAA01001077.1 | 52.90 | full | 178,132 | 178,054 | VXAA01001077.1 Anseranas semipalmata voucher USNM:623235 isolate B10K-DU-001-57 scaffold61, whole genome shotgun sequence | Anseranas semipalmata (magpie goose) |
|
|
| 263 | VXAL01012489.1 | 52.90 | full | 159,662 | 159,740 | VXAL01012489.1 Chauna torquata voucher USNM:614546 isolate B10K-DU-011-36 scaffold1803, whole genome shotgun sequence | Chauna torquata (southern screamer) |
|
|
| 264 | WBMW01001713.1 | 52.90 | full | 633,056 | 632,978 | WBMW01001713.1 Penelope pileata voucher USNM:572502 isolate B10K-DU-001-08 scaffold212, whole genome shotgun sequence | Penelope pileata |
|
|
| 265 | KZ155910.1 | 52.90 | full | 1,743,221 | 1,743,299 | KZ155910.1 Anser cygnoides isolate SCWG-2014 breed Sichuan white goose unplaced genomic scaffold scaffold69, whole genome shotgun sequence | Anser cygnoides (Chinese goose) |
|
|
| 266 | NXHY01000002.1 | 52.90 | full | 19,984,315 | 19,984,237 | NXHY01000002.1 Anser brachyrhynchus strain PFG001 scaffold_1, whole genome shotgun sequence | Anser brachyrhynchus (pink-footed goose) |
|
|
| 267 | VXAU01001621.1 | 52.80 | full | 49,526 | 49,453 | VXAU01001621.1 Formicarius rufipectus voucher USNM:613308 isolate B10K-DU-001-43 scaffold2499, whole genome shotgun sequence | Formicarius rufipectus |
|
|
| 268 | VWZJ01003812.1 | 52.70 | full | 10,887 | 10,810 | VWZJ01003812.1 Hemiprocne comata voucher USNM:607338 isolate B10K-DU-001-23 scaffold275, whole genome shotgun sequence | Hemiprocne comata |
|
|
| 269 | WBNI01000042.1 | 52.40 | full | 1,550,255 | 1,550,330 | WBNI01000042.1 Eolophus roseicapilla voucher USNM:632230 isolate B10K-DU-025-06 scaffold_35, whole genome shotgun sequence | Eolophus roseicapillus (Galah) |
|
|
| 270 | KK684944.1 | 52.30 | full | 7,797 | 7,874 | KK684944.1 Leptosomus discolor isolate BGI_N330 unplaced genomic scaffold scaffold23937, whole genome shotgun sequence | Leptosomus discolor |
|
|
| 271 | KK939260.1 | 52.20 | full | 16,485 | 16,410 | KK939260.1 Nestor notabilis isolate BGI_N333 unplaced genomic scaffold scaffold21995, whole genome shotgun sequence | Nestor notabilis |
|
|
| 272 | VZRH01003229.1 | 52.20 | full | 403,622 | 403,697 | VZRH01003229.1 Urocynchramus pylzowi voucher IZCAS:653 isolate B10K-DU-012-38 scaffold807, whole genome shotgun sequence | Urocynchramus pylzowi |
|
|
| 273 | LR778266.1 | 52.10 | full | 32,499,831 | 32,499,903 | LR778266.1 Coregonus sp. 'balchen' genome assembly, chromosome: 14 | Coregonus sp. 'balchen' |
|
|
| 274 | VZUD01000027.1 | 52.00 | full | 398,949 | 399,024 | VZUD01000027.1 Cepphus grylle isolate OUT-0020 scaffold90_cov51, whole genome shotgun sequence | Cepphus grylle |
|
|
| 275 | JH556605.1 | 51.90 | full | 286,665 | 286,590 | JH556605.1 Melopsittacus undulatus unplaced genomic scaffold budgerigar_v6.3_scf900160277058, whole genome shotgun sequence | Melopsittacus undulatus (budgerigar) |
|
|
| 276 | VYZS01325787.1 | 51.90 | full | 1,341 | 1,266 | VYZS01325787.1 Neodrepanis coruscans voucher FMNH:AVES:345711 isolate B10K-DU-002-79 C19707425, whole genome shotgun sequence | Neodrepanis coruscans (wattled asity) |
|
|
| 277 | KL372813.1 | 51.90 | full | 10,702 | 10,625 | KL372813.1 Apaloderma vittatum isolate BGI_N311 unplaced genomic scaffold scaffold11760, whole genome shotgun sequence | Apaloderma vittatum |
|
|
| 278 | WBMZ01001873.1 | 51.80 | full | 6,118 | 6,193 | WBMZ01001873.1 Atrichornis clamosus isolate B10K-DU-029-61 scaffold7033, whole genome shotgun sequence | Atrichornis clamosus |
|
|
| 279 | SCEB01214181.1 | 51.60 | full | 154,252 | 154,326 | SCEB01214181.1 Acipenser ruthenus isolate WHYD16114868_AA scaffold1113, whole genome shotgun sequence | Acipenser ruthenus (sterlet) |
|
|
| 280 | VWZT01011989.1 | 51.50 | full | 111,487 | 111,562 | VWZT01011989.1 Pachycephala philippinensis voucher USNM:607512 isolate B10K-DU-001-28 scaffold4802, whole genome shotgun sequence | Pachycephala philippinensis |
|
|
| 281 | JH126755.1 | 51.40 | full | 1,684,625 | 1,684,697 | JH126755.1 Latimeria chalumnae unplaced genomic scaffold scaffold00194, whole genome shotgun sequence | Latimeria chalumnae (coelacanth) |
|
|
| 282 | KN126173.1 | 51.40 | full | 2,596,928 | 2,596,851 | KN126173.1 Chaetura pelagica isolate M959 unplaced genomic scaffold scaffold363, whole genome shotgun sequence | Chaetura pelagica (chimney swift) |
|
|
| 283 | VWZS01004484.1 | 51.20 | full | 862,201 | 862,278 | VWZS01004484.1 Rhabdornis inornatus voucher USNM:607544 isolate B10K-DU-001-29 scaffold866, whole genome shotgun sequence | Rhabdornis inornatus |
|
|
| 284 | VZSJ01002315.1 | 51.10 | full | 447,729 | 447,654 | VZSJ01002315.1 Pheucticus melanocephalus isolate OUT-0018 scaffold7275, whole genome shotgun sequence | Pheucticus melanocephalus |
|
|
| 285 | CM028489.1 | 51.10 | full | 7,418,420 | 7,418,498 | CM028489.1 Gallus gallus isolate bGalGal1 chromosome 8, whole genome shotgun sequence | Gallus gallus (chicken) |
|
|
| 286 | KZ860780.1 | 51.10 | full | 391,772 | 391,694 | KZ860780.1 Chrysolophus pictus breed wild golden pheasant unplaced genomic scaffold scaffold95, whole genome shotgun sequence | Chrysolophus pictus (golden pheasant) |
|
|
| 287 | QCWP01000068.1 | 51.10 | full | 643,363 | 643,285 | QCWP01000068.1 Phasianus colchicus isolate SZU-A5-319 scaffold82_cov58, whole genome shotgun sequence | Phasianus colchicus (Ring-necked pheasant) |
|
|
| 288 | QJSC01002381.1 | 51.00 | full | 16,381,208 | 16,381,283 | QJSC01002381.1 Calidris pygmaea strain Meinypilgyno scaf_10611, whole genome shotgun sequence | Eurynorhynchus pygmeus |
|
|
| 289 | VXAK01015759.1 | 51.00 | full | 139,581 | 139,656 | VXAK01015759.1 Arenaria interpres voucher USNM:607650 isolate B10K-DU-005-73 scaffold9472, whole genome shotgun sequence | Arenaria interpres (ruddy turnstone) |
|
|
| 290 | VZRU01010649.1 | 51.00 | full | 11,406 | 11,481 | VZRU01010649.1 Pedionomus torquatus voucher MVMA:2319 isolate B10K-DU-029-80 scaffold4581, whole genome shotgun sequence | Pedionomus torquatus (plains-wanderer) |
|
|
| 291 | VWZA01000395.1 | 50.90 | full | 220,112 | 220,037 | VWZA01000395.1 Oceanites oceanicus voucher LSUMZ:Ornithology:B46846 isolate B10K-CU-031-11 scaffold_388, whole genome shotgun sequence | Oceanites oceanicus |
|
|
| 292 | VYZH01000893.1 | 50.90 | full | 69,708 | 69,783 | VYZH01000893.1 Probosciger aterrimus voucher USNM:615002 isolate B10K-DU-017-47 scaffold_877, whole genome shotgun sequence | Probosciger aterrimus |
|
|
| 293 | VZZT01000158.1 | 50.90 | full | 310,441 | 310,366 | VZZT01000158.1 Fregetta grallaria voucher USNM:615151 isolate B10K-DU-006-09 scaffold_153, whole genome shotgun sequence | Fregetta grallaria |
|
|
| 294 | LR735557.1 | 50.90 | full | 61,524,704 | 61,524,629 | LR735557.1 Sarcophilus harrisii genome assembly, chromosome: 4 | Sarcophilus harrisii (Tasmanian devil) |
|
|
| 295 | CM040764.1 | 50.80 | full | 19,187,833 | 19,187,761 | CM040764.1 Albula goreensis ecotype Florida chromosome 24, whole genome shotgun sequence | Albula goreensis |
|
|
| 296 | VWPQ01004056.1 | 50.80 | full | 67,494 | 67,419 | VWPQ01004056.1 Ceuthmochares aereus voucher LSUMZ:Ornithology:B45261 isolate B10K-CU-031-02 scaffold_4050, whole genome shotgun sequence | Ceuthmochares aereus |
|
|
| 297 | WAAB01002280.1 | 50.80 | full | 868 | 793 | WAAB01002280.1 Piaya cayana voucher USNM:632527 isolate B10K-DU-008-47 scaffold9875, whole genome shotgun sequence | Piaya cayana (squirrel cuckoo) |
|
|
| 298 | QZWQ01000180.1 | 50.60 | full | 1,330,862 | 1,330,940 | QZWQ01000180.1 Pavo cristatus isolate SKPea2016_SI scaffold22_len1268288_cov0, whole genome shotgun sequence | Pavo cristatus (Indian peafowl) |
|
|
| 299 | QZWQ01000213.1 | 50.60 | full | 3,052 | 3,130 | QZWQ01000213.1 Pavo cristatus isolate SKPea2016_SI scaffold259_len197532_cov0_single, whole genome shotgun sequence | Pavo cristatus (Indian peafowl) |
|
|
| 300 | QZWQ01009865.1 | 50.60 | full | 3,177 | 3,255 | QZWQ01009865.1 Pavo cristatus isolate SKPea2016_SI scaffold10117_len7922_cov0_single, whole genome shotgun sequence | Pavo cristatus (Indian peafowl) |
|
Note: this table contains only the first 300 sequences.
Alignment
There are various ways to view or download the seed alignments that we store. You can use a sequence viewer to look at them, or you can look at a plain text version of the sequence in a variety of different formats. More...
View options
You can view Rfam seed alignments in your browser in various ways. Choose the viewer that you want to use and click the "View" button to show the alignment in a pop-up window.
Formatting options
You can view or download Rfam seed alignments in several formats. Check either the "download" button, to save the formatted alignment, or "view", to see it in your browser window, and click "Generate".
Download
Download a gzip-compressed, Stockholm-format file containing the seed alignment for this family. You may find RALEE useful when viewing sequence alignments.
Submit a new alignment
We're happy receive updated seed alignments for new or existing families. Submit your new alignment and we'll take a look.
Secondary structure
This section shows a variety of different secondary structure representations for this family. More...
You can view the secondary structure of the family using the VARNA applet. You can see more information about VARNA iself here.
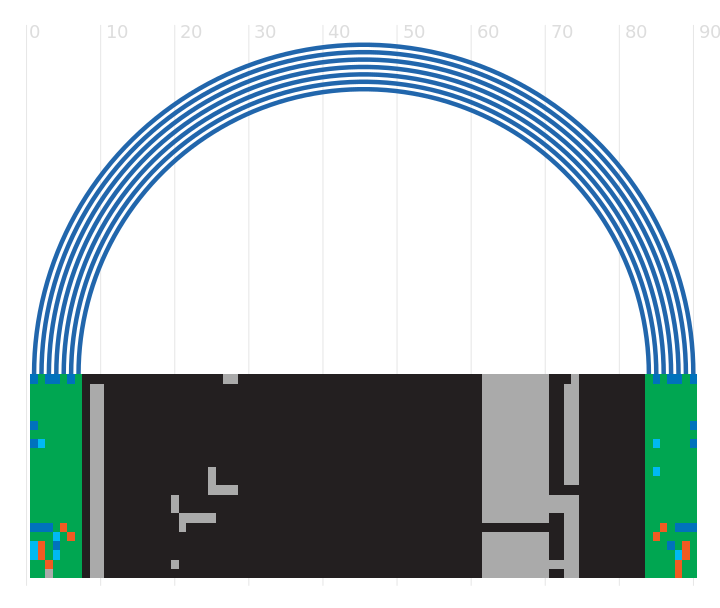
Current Rfam structure
1 out of 7 basepairs are significant at E-value=0.05
R-scape optimised structure
1 out of 1 basepairs are significant at E-value=0.05
- Colours
- Statistically significant basepair with covariation
- 97% conserved nucleotide
- 90% conserved nucleotide
- 75% conserved nucleotide
- 50% conserved nucleotide
- Nucleotides
- R: A or G
- Y: C or U
Tip: The diagrams are interactive:
you can pan and zoom to see more details
or hover over nucleotides and basepairs.
R-scape is a method for testing whether covariation analysis supports the presence of a conserved RNA secondary structure. This page shows R-scape analysis of the secondary structure from the Rfam seed alignment and a new structure with covariation support that is compatible with the same alignment.
To find out more about the method, see the R-scape paper by Rivas et al., 2016. The structures are visualised using R2R.
Species distribution
Sunburst controls
HideLineage
Move your mouse over the main tree to show the lineage of a particular node.
You can move this pane by dragging it.
Weight segments by...
Change the size of the sunburst
Colour assignments
 Archea
Archea
|
 Eukaryota
Eukaryota
|
 Bacteria
Bacteria
|
 Other sequences
Other sequences
|
 Viruses
Viruses
|
 Unclassified
Unclassified
|
 Viroids
Viroids
|
 Unclassified sequence
Unclassified sequence
|
Selections
Click on a node to select that node and its sub-tree.
Clear selection
This visualisation provides a simple graphical representation of the distribution of this family across species. You can find the original interactive tree in the adjacent tab. More...
Tree controls
HideThe tree shows the occurrence of this RNA across different species. More...
Loading...
Please note: for large trees this can take some time. While the tree is loading, you can safely switch away from this tab but if you browse away from the family page entirely, the tree will not be loaded.
Trees
This page displays the predicted phylogenetic tree for the alignment. More...
Note: You can also download the data file for the seed tree.
References
This section shows the database cross-references that we have for this Rfam family.
Literature references
-
Smith CM, Steitz JA; Mol Cell Biol 1998;18:6897-6909. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5'-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. PUBMED:9819378
External database links
| Gene Ontology: | GO:0006396 (RNA processing); GO:0005730 (nucleolus); |
| Sequence Ontology: | SO:0000593 (C_D_box_snoRNA); |
Curation and family details
This section shows the detailed information about the Rfam family. We're happy to receive updated or improved alignments for new or existing families. Submit your new alignment and we'll take a look.
Curation
| Seed source | Griffiths-Jones SR | ||||||
| Structure source | Predicted; PFOLD | ||||||
| Type | Gene; snRNA; snoRNA; CD-box; | ||||||
| Author |
Griffiths-Jones SR
|
||||||
| Alignment details |
|
Model information
| Build commands |
cmbuild -F CM SEED
cmcalibrate --mpi CM
|
| Search command |
cmsearch --cpu 4 --verbose --nohmmonly -T 29.50 -Z 2958934 CM SEQDB
|
| Gathering cutoff | 50.0 |
| Trusted cutoff | 50.0 |
| Noise cutoff | 49.7 |
| Covariance model | Download |
 Loading...
Loading...